Abstract
Ultrasonography (US) is noninvasive and offers real-time, low-cost, and portable imaging that facilitates the rapid and dynamic assessment of musculoskeletal components. Significant technological improvements have contributed to the increasing adoption of US for musculoskeletal assessments, as artificial intelligence (AI)-based computer-aided detection and computer-aided diagnosis are being utilized to improve the quality, efficiency, and cost of US imaging. This review provides an overview of classical machine learning techniques and modern deep learning approaches for musculoskeletal US, with a focus on the key categories of detection and diagnosis of musculoskeletal disorders, predictive analysis with classification and regression, and automated image segmentation. Moreover, we outline challenges and a range of opportunities for AI in musculoskeletal US practice.
Go to : 

Introduction
Ultrasound (US) imaging is a useful diagnostic tool for the examination of the musculoskeletal system. It provides real-time imaging without ionizing radiation and is noninvasive. It has also gained popularity for the dynamic imaging of small structures and the evaluation of the ligaments, muscle and tendons, superficial tumors, and peripheral nerves [1]. Progressive advances in US, including refined transducer technology, power Doppler sonography, and real-time US elastography (EUS), have expanded its clinical applications in the field of musculoskeletal imaging. Additionally, new capabilities that enhance spatial resolution and image quality, such as speckle reduction, video capturing, harmonic tissue imaging, compound imaging, and panoramic imaging, have emerged following revolutionary innovations in computing power and algorithms [2]. For example, EUS facilitates the accurate detection of subclinical changes in the muscles and tendons by leveraging the mechanical properties of musculoskeletal tissue for early diagnosis and therapy monitoring [3].
Within the last decade, classical computer-aided diagnosis (CADx) systems emerged as analytic tools that incorporated a selected set of quantitative features (e.g., first-, second- and higher-order statistical features) to detect abnormal regions in US images accurately. Various machine learning (ML) techniques have been developed to support high-performance computer-aided detection (CADe) or CADx systems for the detection of clinically significant regions, automated segmentation, and classification based on extracted radiological features [4,5]. The image processing techniques used with ML algorithms automate the diagnostic process of detection and characterization for numerous diseases. However, they are limited by their inability to generalize different parameters and high dependence on the settings of the US scanner and image acquisition system. For musculoskeletal US imaging, the integration of conventional ML and CADe or CADx techniques has also been limited by small, thin, narrow, or curved anatomical structures such as extensor digitorum tendons, tiny ligaments or retinacula of the hand, and small nerve branches. These structures may have thicknesses ranging between 0.1 and 0.6 mm [6], making them prone to segmentation and classification errors. Additionally, although US has been explored for musculoskeletal imaging, the curation of US musculoskeletal data remains challenging. Concurrently, the identification of the sonographic appearance of specific anatomical structures remains suboptimal and is still evolving [7], and relevant datasets of musculoskeletal US images with expert annotations are limited.
Over the years, deep learning (DL) has become a notable subfield of artificial intelligence (AI) for high-quality image interpretation and acquisition, and offers support to health professionals for objective and accurate US image analysis [8]. DL in musculoskeletal US is receiving significant attention for automated feature engineering with deep neural networks (DNNs). DNNs are expected to be pivotal in the development of next-generation cutting-edge US imaging systems, exploiting the intrinsic complexities of the anatomical structures and tissues to drive effective triaging and rapid diagnosis. This paper provides an overview of definitions and reviews the recent literature on applications of AI in musculoskeletal US. It also evaluates the future of AI in musculoskeletal US imaging, focusing on key categories of AI-based US image enhancement, classification, detection, and automated segmentation. This article aims to provide musculoskeletal professionals with an understanding of AI technology and to outline the implications of its integration in musculoskeletal US for real-world clinical practice.
Go to : 

What Is AI?
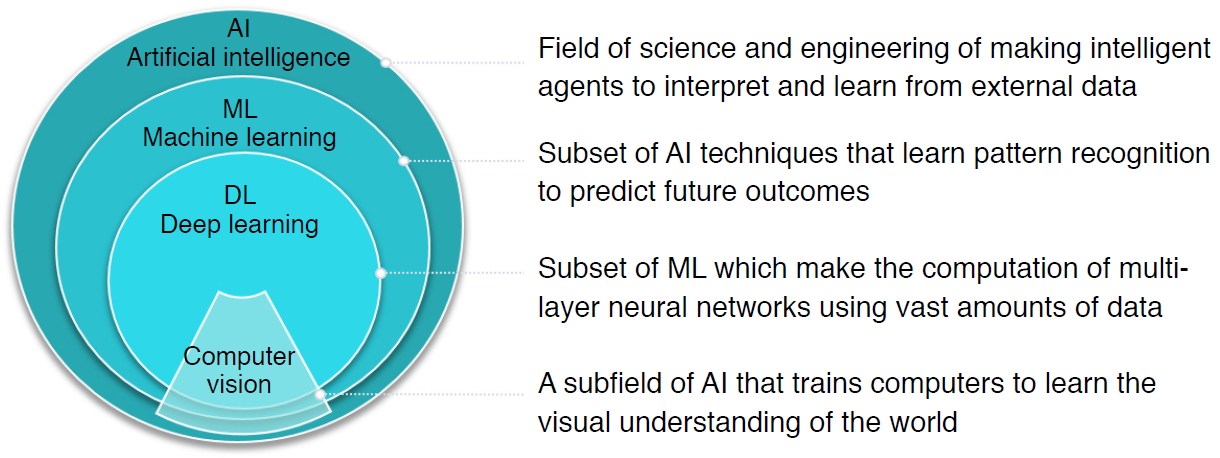
AI is defined as a field of science and engineering that seeks to create intelligent machines to interpret and learn from external data and achieve specific objectives, including natural language processing, robotics, and ML. As a subset of AI, ML is a powerful set of computational tools that have incredible pattern-recognizing abilities, enabling them to automate the reasoning processes of experts (Fig. 1).
Go to : 

ML: Feature Extraction and Classification Algorithms
Prior to the explosive growth of DL, ML-based strategies that trained models on descriptive patterns obtained from rules of human inference were dominant [4]. Researchers manually transformed raw data into features, followed by the selection of the best features (e.g., intensity histograms, texture-based features, geometric features, and morphological features). Subsequently, traditional ML classification algorithms (e.g., random forest and support vector machine [SVM] algorithms) were applied to the extracted features [9] (Fig. 2A). A comprehensive review of ML studies shows numerous high-quality techniques to evaluate musculoskeletal disorders based on US images (Table 1) [10-15]. However, a major challenge facing these approaches is that feature selection heavily relies on statistical insights and domain knowledge, and this limitation initiated a paradigm shift from manual feature engineering to DL architectural design.
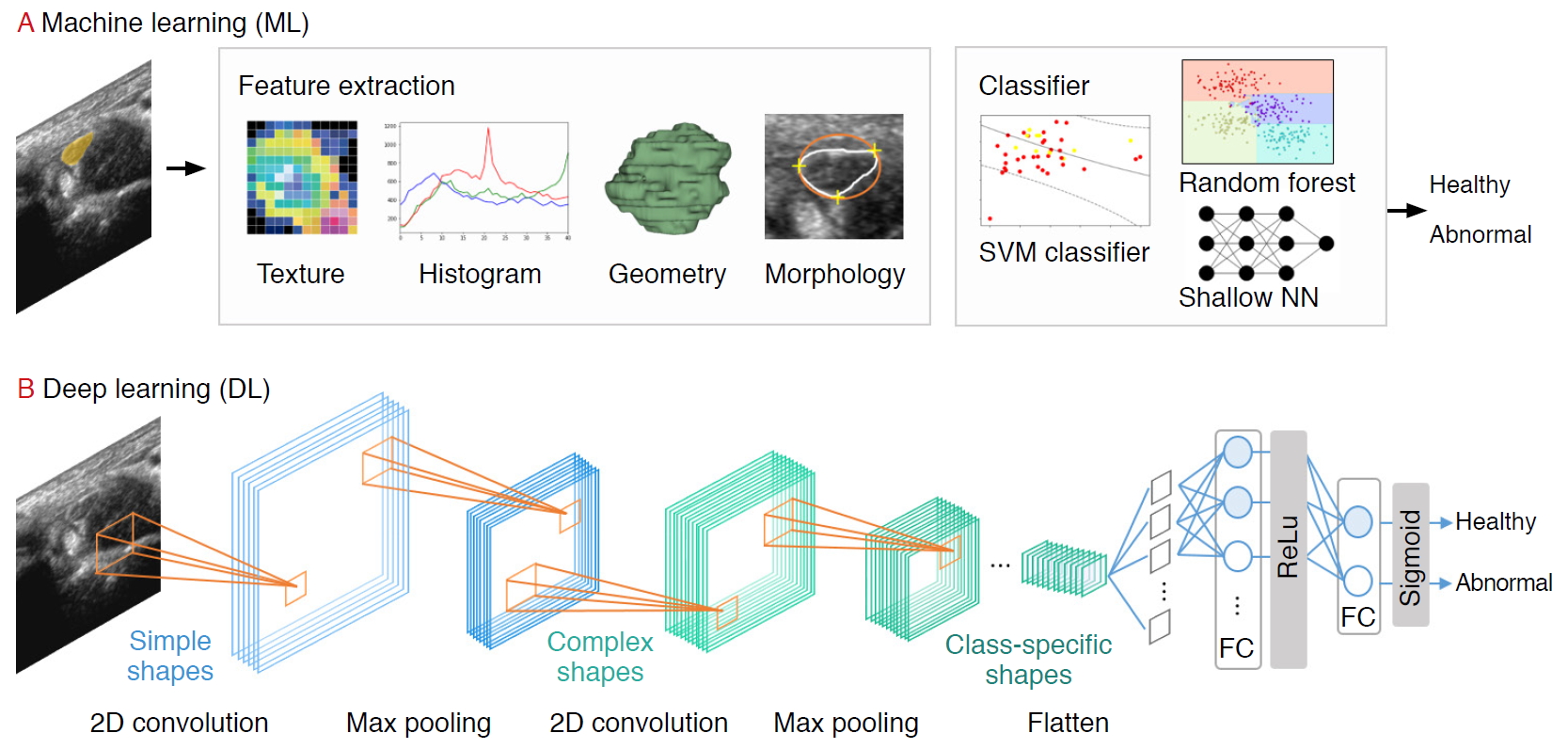 | Fig. 2.A pipeline of ultrasound research using machine learning (ML) and deep learning (DL).For the ML pipeline, the practitioner extracts the features (e.g., texture, histogram, geometry, morphology) manually before feeding it into the classification model. In the DL pipeline, features are extracted automatically using convolutional filters and pooling. SVM, support vector machine. |
Table 1.
Overview of machine learning algorithms and applications used in musculoskeletal ultrasound imaging
| Algorithm | Advantage | Limitation | Example application in musculoskeletal ultrasonography |
|---|---|---|---|
| Logistic regression | Provides probabilistic interpretation of model parameters | Only used to predict discrete function | - |
| Quick model update for incorporating new data | Sensitive to outliers | ||
| K-nearest neighbors | Nonparametric model | Time-consuming and computationally expensive | Nerve identification [10] |
| Used both for classification and regression problems | Number of neighbors must be defined in advance | ||
| Low interpretability | |||
| Naïve Bayes | Suitable for relatively small datasets | Classes must be mutually exclusive | - |
| Handles both binary and multi-class classification problems | Presence of dependency between attributes results in loss of accuracy | ||
| Fast application and high computational efficiency | Assumptions such as the normal distribution might be invalid | ||
| Support vector machines | Good prediction performance in different tasks | Have "black box" characteristics | Lumbar spine classification [11] |
| Can handle multiple feature spaces | Sensitive to manual parameter tuning and kernel choice | Synovitis grading [12] | |
| Nerve identification [10] | |||
| Decision trees | Perform in datasets with large number of features | Only axis-aligned rectangle splits. | Nerve identification [10] |
| Few parameter tuning | Inadequate for regression and continuous value prediction problems | ||
| High representational power and easy to interpret | Mistake in higher labels cause errors in subtrees | ||
| Random forest | Provide estimates of variable or attribute importance in the classification | Complex and computationally expensive | Myositis classification [13] |
| Ensemble-based classifications shows relatively good performance | Number of base classifiers needs to be defined | Hip 2-D US adequacy classification [14] | |
| Overfitting has been observed for noisy data | |||
| Neural networks | Direct image processing | Have "black box" characteristics | Nerve identification [10] |
| Can map complex nonlinear relationships between dependent and independent variables | Have to fine-tune many parameters | ||
| Require a large well-annotated dataset to achieve good performance | |||
| K-means | Can process large datasets | Number of clusters must be defined | Nerve localization [15] |
| Algorithm that is simple to understand and implement |
Go to : 

DL: Convolutional Neural Networks
DL, based on increasing the number of hidden neural network layers, revolutionized the field of end-to-end learning by bypassing the hand-crafted engineering stages that characterize ML pipelines (Fig. 2A) [4]. While conventional radiological assessments are often based on radiologists’ knowledge and experience, DNNs automatically recognize patterns from data and achieve remarkable performance in various applications in radiology [16,17], dermatology [18], and ophthalmology [19], among others.
Several DL applications in radiology are supervised, and most of them are based on convolutional neural networks (CNNs). CNNs comprise a sequence of hidden layers, each responding to unique features that transform images into output class scores (Fig. 2B). CNNs are biologically-inspired neural networks that mimic the physiology of the visual cortex by responding differently to specific features [20]. Simple cells in the visual cortex are the most specific, as they detect lines, edges, and corners in a visual field. Complex features such as colors, shapes, and orientations are captured by complex cells, which show more spatial invariance by pooling the outputs of simple cells. Similar to human visual perception, which is regulated by the visual cortex, two main characteristics make CNNs optimum for image classification: the increasing shape "selectivity" and "the invariance" of the visual representation through feedforward connections.
A CNN is composed of a series of three layers: a convolutional (CONV) layer, followed by a pooling layer, and finally, a fully connected layer (Fig. 2B). The CONV layer forms the basis of the CNN, containing a set of filters with parameters that need to be learned. During the forward pass, the input is convolved by several filters to compute two-dimensional (2D) activation maps of every spatial region. These activations are processed layer by layer to extract high-level features. Repeated convolution of the image results in a map of activations, called feature maps, which represents the location and strength of unique features, pixels, and characters invariant to translation. The pooling layer performs a down-sampling of the spatial dimension and reduces the spatial sizes of the representations. Thus, the number of parameters to be learned and computational complexity is decreased. The fully connected layer maintains full connectivity between the neurons of each preceding and succeeding layer. The CONV and pooling layers perform feature extractions of the given image, and the fully connected layer acts as a classifier that discriminates based on the high-level representation of images.
[cont.]


Không có nhận xét nào :
Đăng nhận xét