AI in Musculoskeletal US Imaging
The latest improvements in US imaging technology have been linked to improved accuracy in the diagnosis of musculoskeletal disorders. However, the dependence on subjective assessments of displayed images and the variability in image acquisition and equipment used across studies [21] have delayed the widespread of US compared to that of magnetic resonance imaging (MRI). To help overcome these problems, as well as the ambiguity with which musculoskeletal disorders may present on US imaging, CADe/CADx has become a major solution in radiology.
CADe/CADx systems provide quantitative analysis and a complimentary opinion that supports radiologists in making accurate and consistent image assessments quickly. The early versions of CADe/CADx systems in musculoskeletal US imaging were based on hand-crafted ML engineering with multiple processing stages: (1) image preprocessing with segmentation and region of interest (ROI) designation, (2) feature extraction and selection, and (3) classification based on the selected features. However, classical CADe/CADx systems suffer from various algorithmic limitations related to data gathering, preprocessing, filtering, and generalization. Evaluations are also restricted due to bias and variance caused by finite samples collected from different medical centers. DL-based US CADe/CADx systems have been proposed for next-generation AI-powered radiology because trained models can be robust and generalizable given the availability of big data in digitalized radiology. Currently, the most notable application of AI in musculoskeletal imaging is pattern recognition and image classification. Unlike the applications of AI of image reconstruction, enhancement, or synthesis for other modalities (e.g., MRI or computed tomography), AI applied to musculoskeletal US focuses mainly on diagnosis, classification, or segmentation (Fig. 3).
 | Fig. 3.Schematic of artificial intelligence (AI)-based machine learning and deep learning applications in musculoskeletal ultrasound imaging of AI-based diagnosis and classification and AI-based automated image segmentation.SVM, support vector machine; KNN, k-Nearest Neighbor; ROI, region of interest. |
Go to : 

Automatic Diagnosis and Detection in Musculoskeletal US
Muscle Disorders
Skeletal muscle US is a point-of-care technique for visualizing skeletal muscle structure, fatty atrophy, movement, and function in real-time. The portability and high spatial resolution of US make it an ideal imaging modality for the detection and diagnosis of muscle injuries, myopathy, or myositis [22]. However, due to variations in operator-dependent techniques and the intra- and inter-reader variability in the qualitative assessment of muscle echogenicity [23,24], computer-aided quantitative methods for the detection of muscle pathology and identification of structural abnormalities of muscle have been studied.
In earlier studies of quantitative muscle US, muscle echo intensity were obtained by grayscale analysis of regions of interest using numerous texture descriptors for muscle characterization [25-27]. To identify Duchenne muscular dystrophy (DMD), echo intensity, and muscle thickness were quantified to distinguish myopathic muscles with increased echogenicity [28]. To detect disease progression in DMD patients, the grayscale level and quantitative backscatter analysis have shown to be more sensitive than functional assessments [29]. To further characterize different muscle types in US images according to sex, different combinations of first-order and higher-order texture descriptors were shown to be useful for muscle disorders [30].
Further developments have involved comparative analyses of ML and DL applied to muscle US images to automatically or semi-automatically differentiate among inclusion body myositis, polymyositis, dermatomyositis, and normal presentations [13]. The DL strategy improved accuracy in all scenarios compared to conventional ML algorithms (random forest), and this supports the notion that manually engineered features, while useful, may facilitate suboptimal diagnosis and are inadequate for a full characterization of disease complexity compared to automatically generated feature sets from DL models.
In addition to the detection and classification of muscle pathology, ML and DL approaches have been applied to the objective estimation of muscle fiber orientations from B-mode US images to predict a continuous output. Deep residual networks and deconvolutional CNNs have made better predictions or estimations of full-spatial-resolution muscle fibers than the wavelet method and CNNs (Fig. 4) [31,32]. These objective measurements of muscular length change, thickness, and tendon length are useful for evaluating muscles. They are also useful for understanding, diagnosing, monitoring, and treating muscular disorders.
 | Fig. 4.Deep convolutional neural network (DCNN)-based fiber orientation.A. A representation of DCNN predictions for fiber orientation is given. A fiber orientation heatmap is shown in the top image, and a line trace representation overlaid on the ultrasound image is shown in the bottom image. CNN, convolutional neural network. B. The temporal variation in fiber orientation traces of maximum voluntary contraction (starting at 0 second and ending at 2.2 seconds) is given. Reprinted from Cunningham et al. J Imaging 2018;4:29, according to the Creative Commons license. |
Go to : 

Diagnosis of Hip Dysplasia
Developmental dysplasia of the hip (DDH) is a developmental deformity of the hip joint characterized by anatomical abnormalities between the head of the femur and the acetabulum [33]. Detecting DDH as early as possible is crucial to avoid the development of residual hip dysplasia or hip osteoarthritis (OA). However, the current US diagnosis of DDH using 2D US is limited by low interobserver reliability [34] and workflow timing of the US evaluation [35]. Thus, AI-based techniques are preferred for the automated interpretation of 2D and three-dimensional (3D) hip US images. The application of AI facilitates a quick estimation of conventional parameters (e.g., alpha angle, acetabular contact angle [ACA]) to provide higher diagnostic accuracy and to minimize interobserver variability with reduced imaging time.
To achieve accurate and rapid semi-automatic delineation of the acetabulum, a semiautomated segmentation technique was used to generate 3D acetabular surface models interpolated from optimal paths passing through user-defined seed points [36]. The ACA delineated from the segmented 3D surface was used to classify the acetabulum as normal, borderline, or dysplastic. Additionally, a fully automated DDH diagnostic technique was proposed, using DNN segmentation with an adversarial network to automatically segment landmarks to estimate Graf's alpha angle in US of infants’ hips [37]. Hareendranathan et al. [38] presented a novel segmentation pipeline, in which US images are first segmented into multiple clusters using a simple linear iterative clustering algorithm. Subsequently, a CNN is trained to classify each segmented cluster, outputting a high-probability region defining the acetabular contour.
Due to advances in segmentation with deep architectures [39-41] and high-level semantic segmentation through complete scene understanding or attentional supervision, it is highly anticipated that DL techniques will replace AI-based applications for DDH assessment. Additionally, the segmentation of 3D inputs using different DL networks [42] to measure the ACA using 3D US may provide a more reliable diagnosis with lower inter-scan variability and shorter processing time.
Go to : 

Automatic Regression and Classification of Musculoskeletal US
Synovitis Assessment
In recent years, US has been used in clinical practice to assist in the assessment and the characterization of patients with synovial proliferative disorders such as inflammatory arthritis, particularly rheumatoid arthritis. The recently published OMERACT-EULAR Synovitis Scoring (OESS) system standardized US scanning for grading synovitis, representing a significant step toward monitoring inflammatory arthritis disease activity [43,44]. Different automated techniques for detection of the synovitis region, quantification of the synovium based on segmentation, and regression models to grade the severity of synovitis have been actively studied.
Synovitis scoring using AI-based computerized techniques may reduce the existing discrepancies between readers [45]. However, designing an automated framework for the quantitative assessment of synovitis in US images is challenging since it is necessary to differentiate synovitis from extra-articular structures, including skin borders and bone regions, under conditions of blurred margins and inhomogeneous echogenicity. The most widely used design of an automated framework for synovitis assessment follows the pipeline of (1) skin border selection, (2) bone location, and (3) synovitis region segmentation. An automated tool for estimation and grading of the synovitis is designed to detect the skin, bone, and joint synovitis through image binarization and image statistics thresholding [46] or in combination with ML classification (SVMs) (Fig. 5) [12].
 | Fig. 5.Example of synovitis area detection with a machine learning-based pipeline, as suggested by Mielnik et al. [12].A-D. The detection of synovitis in the proximal interphalangeal joint (A), detection of synovitis in the metacarpophalangeal joint (B), example of underestimated region of synovitis (C), example of error in synovial hypertrophy detection (D) are shown. Reprinted from Mielnik et al. Ultrasound Med Biol 2018;44:489-494, Copyright (2020), with permission from Elsevier [12]. |
Recently, DL has shown the potential for joint detection and synovitis grading. A hybrid of image processing and an intensity-based algorithm has been proposed for skin border segmentation, while a connectivity algorithm has been proposed for bone region segmentation [47], to grade synovitis. Deep CNNs (DCNNs) have demonstrated the potential to serve as a feasible method for classifying disease activity according to the OESS system. Two DCNN architectures (VGG-16 for classifying healthy or diseased, and Inception-V3 for classifying OESS scores) were applied to Doppler US images, and they achieved high accuracy for low and high-level classification (area under the curve, 0.93) and full-scale OESS classification (quadratically weighted kappa, 0.84) [48]. The applications of DCNNs to the direct quantification of synovitis from entire US images are promising; however, to support an automated DL-based diagnosis, visualization methods such as attention maps that highlight the radiological areas of interest [49] must be integrated into US applications to maintain transparency in the decision process. Given the technological process, dynamic models (e.g., recurrent neural networks) predicting disease progression or quantitative assessment of 3D synovial proliferation and synovitis can be expected.
Go to : 

Spine Level Analysis and Identification
US-guided interventions are gaining popularity for facilitating vascular access, peripheral nerve blocks, and neuraxial anesthesia [50]. Unfortunately, the visualization of spinal US images remains indirect due to inherent speckle noise and the acoustic shadow cast by bone surfaces that hide key anatomical sites, making spinal US technically challenging for a novice who has received limited training in reading US images. Therefore, computer processing algorithms and ML methods have been applied to identify the needle puncture site to aid in US image interpretation. For this, template matching techniques such as lamina and ligamentum flavum (LF) detection [51], midline detection [11], and signatory features detection [52] have been used to identify the skin-to-LF depth and bone structures (Fig. 6). However, the aforementioned template matching-based approaches use a static template stored from a subset of finite subjects for target identification that cannot cover the complete inter-patient variability of spine structures and surroundings.
 | Fig. 6.Image identification for lumbar ultrasound image.The pipeline proposed by Yu et al. [11] consists of a feature extraction method to extract important anatomic features and midline detection and classification stage for interspinous region identification. SVM, support vector machine. Reprinted from Yu et al. Ultrasound Med Biol 2015;41:2677-2689, Copyright (2020), with permission from Elsevier [11]. |
Recently, ML feature extraction and neural network classification techniques were proposed to automate the identification of the optimum plane for epidural steroid and facet joint injections [53]. A method for vertebral localization in the operation room by registering the spinous process shape from radiograph spinous annotations and U-Net segmentation of sagittal US images has been proposed to provide tracker-free 2D US imaging [54]. To visualize the spinal anatomy while performing needle insertion, the SLIDE system [55] has been proposed to classify three characteristic transverse planes, namely the sacrum, intervertebral gaps, and vertebral bones, in real-time. Without the need for predefined features, SLIDE utilizes transfer learning and four different DCNN architectures to learn features of the spine entirely, and a state machine was developed to accurately identify transitions between the planes.
Go to : 

Automated US Image Segmentation Techniques
Nerve Localization and Segmentation
US is the primary diagnostic imaging modality for suspected peripheral neuropathy, particularly when neurological examinations are inconclusive. Nerve conduction velocity or electromyography are commonly analyzed, and morphometric parameters (e.g., cross-sectional area [CSA]) and quantitative measurements of sonographic elastography of peripheral nerves can reflect degrees of peripheral neuropathy. The quantitative evaluation of peripheral nerves using US or sonographic elastography is proceeded by observation of the CSA or assessment of a manually measured ROI. Thus, there is increasing demand for automatic segmentation to extract the ROI of the abnormal signal of the peripheral nerve to reduce the burden of time-consuming and labor-intensive manual measurements. Fully- or semi-automatic AI-based segmentation has shown potential benefits for the segmentation of peripheral nerves due to their near-instantaneous assessment, cost-effectiveness, and high reproducibility.
To detect nerve regions to assist in US segmentation, the majority of classical AI methods are based on four main steps: (1) despeckle filtering, (2) template-based ROI detection, (3) feature-based nerve region classification, and (4) segmentation (Fig. 7). The method proposed by Hadjerci et al. [56] uses the median binary pattern and the Gabor filter to the separated hyperechoic tissues, followed by SVM extraction of the nerve region. Furthermore, the authors reported a comparative study of nerve segmentation with a quantitative performance evaluation using 11 despeckling filters, six statistical feature extraction methods, filter- and wrapper-based feature selection, and five ML-based classifiers [10]. However, these classical feature representations require extensive reformulation of numerous frameworks, statistical insights, and expert domain knowledge, and optimality is not guaranteed.
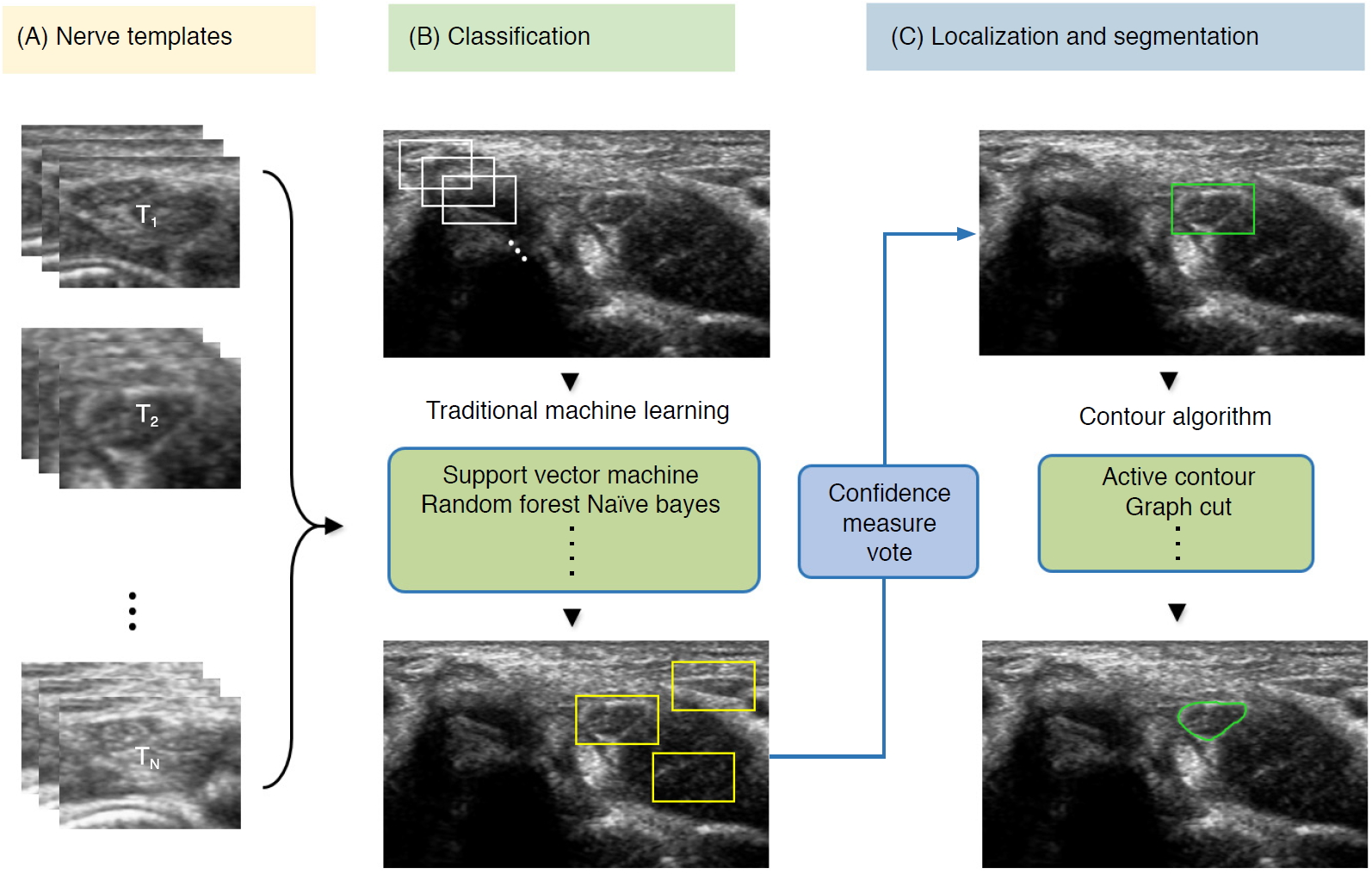 | Fig. 7.Conventional machine learning-based segmentation scheme of nerve ultrasonography.Sliding window template-based classification is applied to generate candidate regions of interest. The nerve region is localized based on a confidence measure vote, and segmentation is applied to obtain nerve boundaries. |
To alleviate the time inefficiency of human interventions and to bypass the intermediate stages in conventional ML-based pipeline designs, DL-based segmentation (U-Net architecture) has been applied to identify musculocutaneous, median, ulnar, and radial nerves [57], as well as femoral nerve blocks in US images [58]. DCNN-based nerve segmentation with variants inspired by the original U-Net architecture was applied [59,60] to NERVE datasets (brachial plexus segmentation in US images, available at https://www.kaggle.com/c/ultrasound-nerve-segmentation). Weng et al. [61] employed neural architecture search, which used autoML algorithms that returned the best neural network through the sampling of building blocks to create an end-to-end structure, and achieved encouraging results (mean intersection over union, 0.992; Dice similarity coefficient, 0.881) compared to U-Net segmentation. Additionally, to precisely locate the position while alleviating the resolution reduction of the brachial plexus, Liu et al. [62] developed a deep adversarial network and used dilated convolution to incorporate the global anatomical contextual cues and organ elastic deformation. Since some nerve regions have small areas with indistinguishable characterizations on ultrasonographic images, different DL techniques that optimize the echotexture, incorporate global semantic context, and achieve computationally efficient real-time segmentation [63] may eventually be developed to reduce failed detection of false-positive findings.
Cartilage Segmentation
Knee OA is a common joint disease among older adults, and its prevalence has been increasing; symptomatic OA occurred 10% in men and 13% in women aged 60 years or older [64]. Even though radiography and MRI are standard imaging modalities in clinical practice for the diagnosis of OA, ultrasonography could become a complementary modality for triaging cartilage abnormalities [65] with benefits of non-invasiveness, availability, relative affordability, and safety, as it is performed without ionizing radiation. Therefore, fully automated measurements of cartilage thickness using segmentation-based techniques have been proposed for real-time imaging for knee OA diagnosis and monitoring.
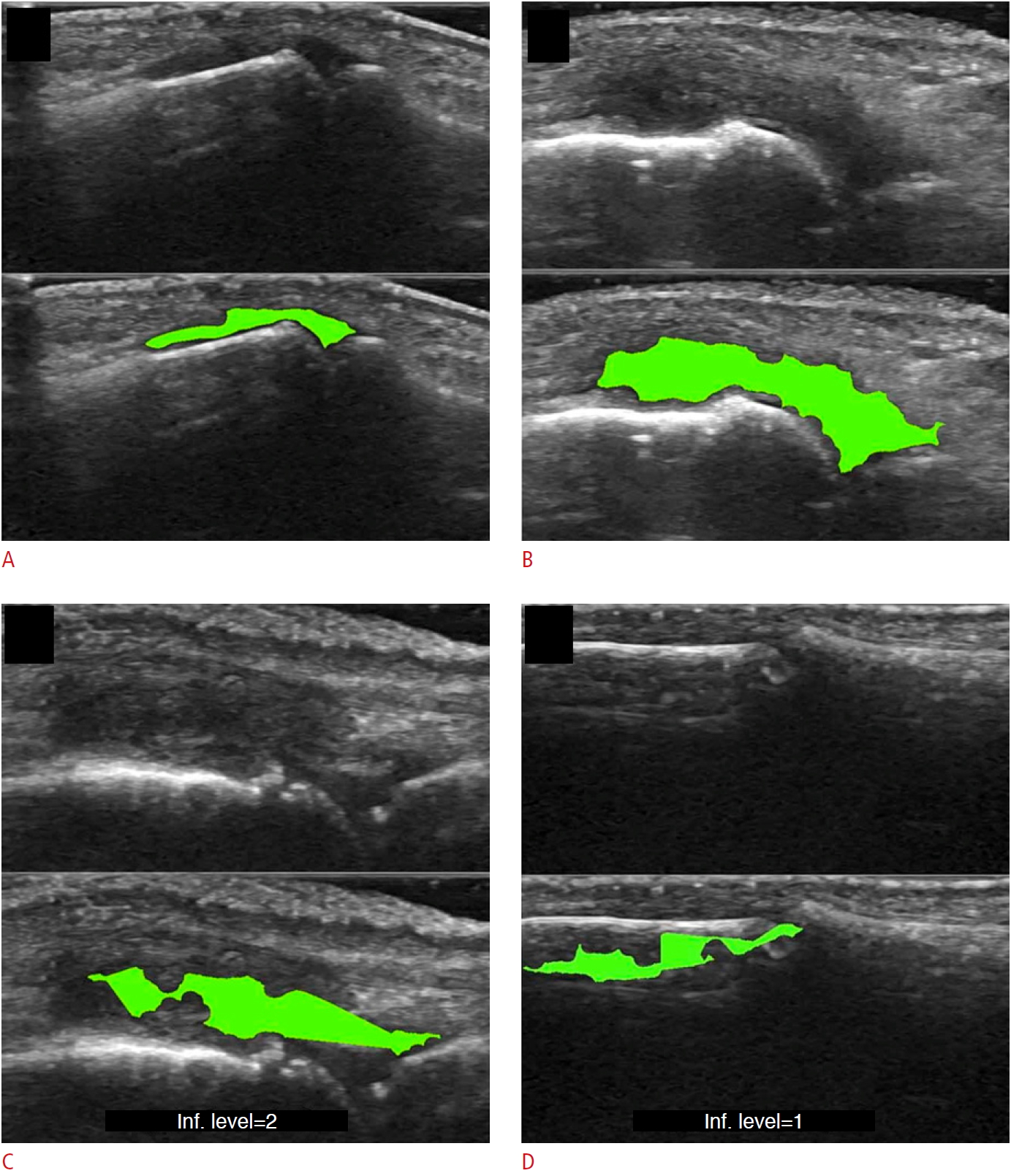
Existing AI-based assessments of cartilage are mainly based on image enhancement and knee cartilage segmentation to represent cartilage thickness with quantitative measures. Hossain et al. [66] used a histogram equalization method of multipurpose beta optimized recursive bi-histogram equalizations to achieve the optimum values of contrast and brightness and to preserve details in the enhancement process of US knee cartilage images. To obtain the true thickness between soft tissue cartilage interface and cartilage-bone interface, quantitative segmentation of the monotonous hypoechoic band was obtained using a locally statistical level set method [67]. Similarly, automated knee-bone surface localization was used to obtain seed points for semi-automatic segmentation methods (random walker, watershed, and graph-cut algorithms), and distance maps are applied to obtain mean cartilage thickness [68]. Recently, the U-Net method has been applied for cartilage segmentation in dynamic, volumetric US images to avoid collision and touching between surgical instruments and anatomical areas during surgery [69] (Fig. 8). Through advances in state-of-art instance segmentation DL models [63], US integrated with DL segmentation might be used to assess of athletic injuries [70], providing accurate real-time feedback.
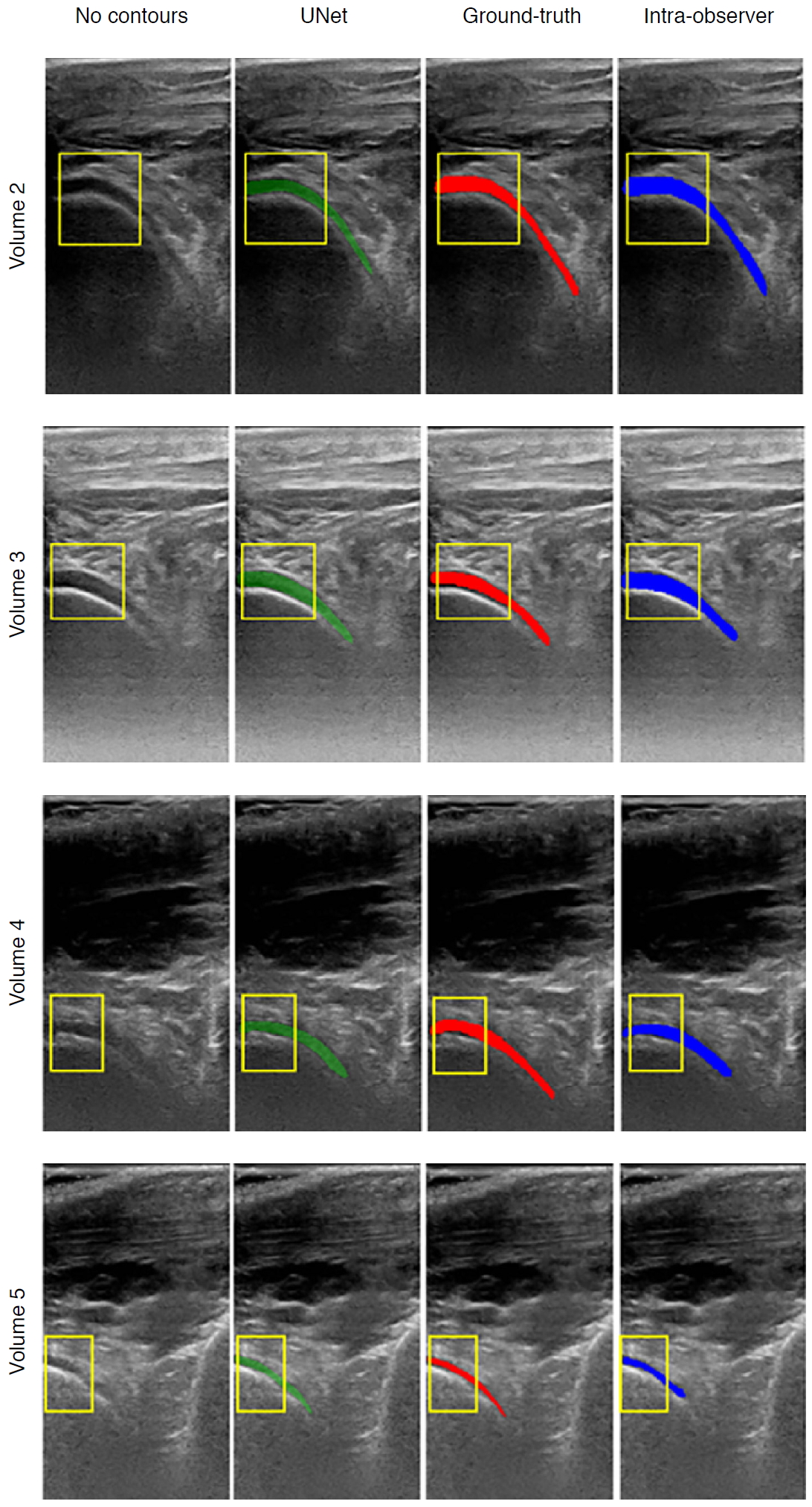 | Fig. 8.Examples of cartilage segmentation based on a U-Net architecture [69].The first column shows examples of images and image regions (yellow box) selected. For each US image in the figure, the segmentations produced by the U-Net (green), by the expert during the ground-truth creation (red), and the intraobserver test (blue) are shown. Reprinted from Antico et al. Ultrasound Med Biol 2020;46:422-435, Copyright (2020), with permission from Elsevier [69]. |
Go to : 

Challenges and Future Perspectives of AI-Based Musculoskeletal US
Although AI-based musculoskeletal US has shown great potential in overcoming high variability and operator dependency, several limitations must be acknowledged. First, there is a discrepancy between 2D imaging, which is widely utilized in radiology clinics, and 3D imaging. Due to the complexity of musculoskeletal structures and various joints, image preprocessing techniques such as rigid or non-rigid image registration are required for the large-scale application of DL for US. Even for US experts, diagnosis based on 2D US is challenging without a comprehensive understanding of functional anatomy. It is challenging to reproduce and localize the thin 2D US image planes, which is disadvantageous for building a large, standardized medical image dataset. Recent AI-based 3D US imaging techniques may overcome the limitations of 2D US [71,72]. Therefore, the strategies for 3D medical US reconstruction, visualization, and segmentation are promising.
Another challenge is that artifacts, which are frequently encountered, can be mistaken for pathology; furthermore, artifacts can occur together with abnormal conditions in both grayscale and Doppler imaging [73]. Careful curation by healthcare professionals and quality assessment of US training data should be considered beforehand, and appropriate preprocessing and normalization are needed to ensure that the artifacts do not affect AI-based model predictions. Artifact reduction and simultaneous preservation of high resolution via generative adversarial networks or automated quality assessment models may be useful for musculoskeletal US imaging.
Finally, image variability due to motion and differences among US machines and transducers constitutes another fundamental limitation hindering the wide adoption of AI-based musculoskeletal ultrasonography. The sonographer's preferred adjustments and further optimization (e.g., different colormaps for grayscale display, dynamic range, edge enhancements, gamma correction, focal depth) lead to additional high variability and randomness, which limit the accuracy and reproducibility of AI models. Additionally, high-quality musculoskeletal annotations are elusive due to the general paucity of expert musculoskeletal radiologists. Therefore, standardized equipment settings, the use of recent image preprocessing software, and open-source datasets that contain extensive collections of annotations based on a high degree of expert consensus may facilitate the better performance of AI-based musculoskeletal US applications and their increased adoption.
Go to : 

Conclusion
Within the last decade, AI-based musculoskeletal imaging has progressed step by step toward enhancing anatomical structure visualization and automating quantitative measurements. Recent studies on AI-based musculoskeletal US have suggested that DL techniques may become next-generation diagnostic tools for monitoring the condition of joints, bones, cartilage, ligaments, and muscles. The image recognition capability of DL may provide sonographers with real-time diagnostic and decision support. By providing high-quality grayscale images, assessing the appropriateness of US images, and ensuring consistency, AI-based musculoskeletal imaging may facilitate higher-quality patient care.
Go to : 

Không có nhận xét nào :
Đăng nhận xét